Share this
Authors
Naho Akiyama, Kensuke Ishiguro, Takeshi Yokoyama, Kenjyo Miyauchi, Asuteka Nagao, Mikako Shirouzu, Tsutomu Suzuki
Abstract
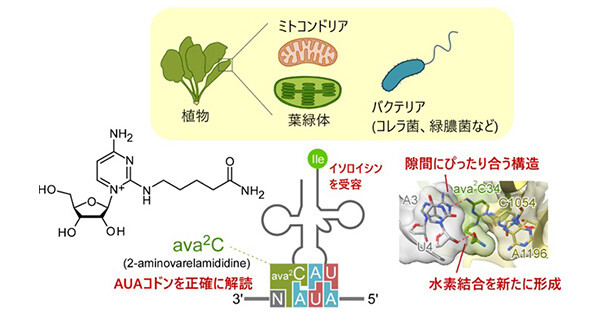
The anticodon modifications of transfer RNAs (tRNAs) finetune the codon recognition on the ribosome for accurate translation. Bacteria and archaea utilize the modified cytidines, lysidine (L) and agmatidine (agm2C), respectively, in the anticodon of tRNAIle to decipher AUA codon. L and agm2C contain long side chains with polar termini, but their functions remain elusive. Here we report the cryogenic electron microscopy structures of tRNAsIle recognizing the AUA codon on the ribosome. Both modifications interact with the third adenine of the codon via a unique C–A geometry. The side chains extend toward 3′ direction of the mRNA, and the polar termini form hydrogen bonds with 2′-OH of the residue 3′-adjacent to the AUA codon. Biochemical analyses demonstrated that AUA decoding is facilitated by the additional interaction between the polar termini of the modified cytidines and 2′-OH of the fourth mRNA residue. We also visualized cyclic N6-threonylcarbamoyladenosine (ct6A), another tRNA modification, and revealed a molecular basis how ct6A contributes to efficient decoding.
Nature Structural & Molecular Biology: https://www.nature.com/articles/s41594-024-01238-1
These Related Stories